Skip to content
2025:
2024:
Wang, Y., Theodore, M. , Xing, Z., Narsaria, U., Yu, Z., Zeng, L., and Zhang, J. , “Structural mechanisms of Tad pilus assembly and its interaction with an RNA virus “, Science Advances . 2024 May 3; 10(18), doi: 10.1126/sciadv.adl4450
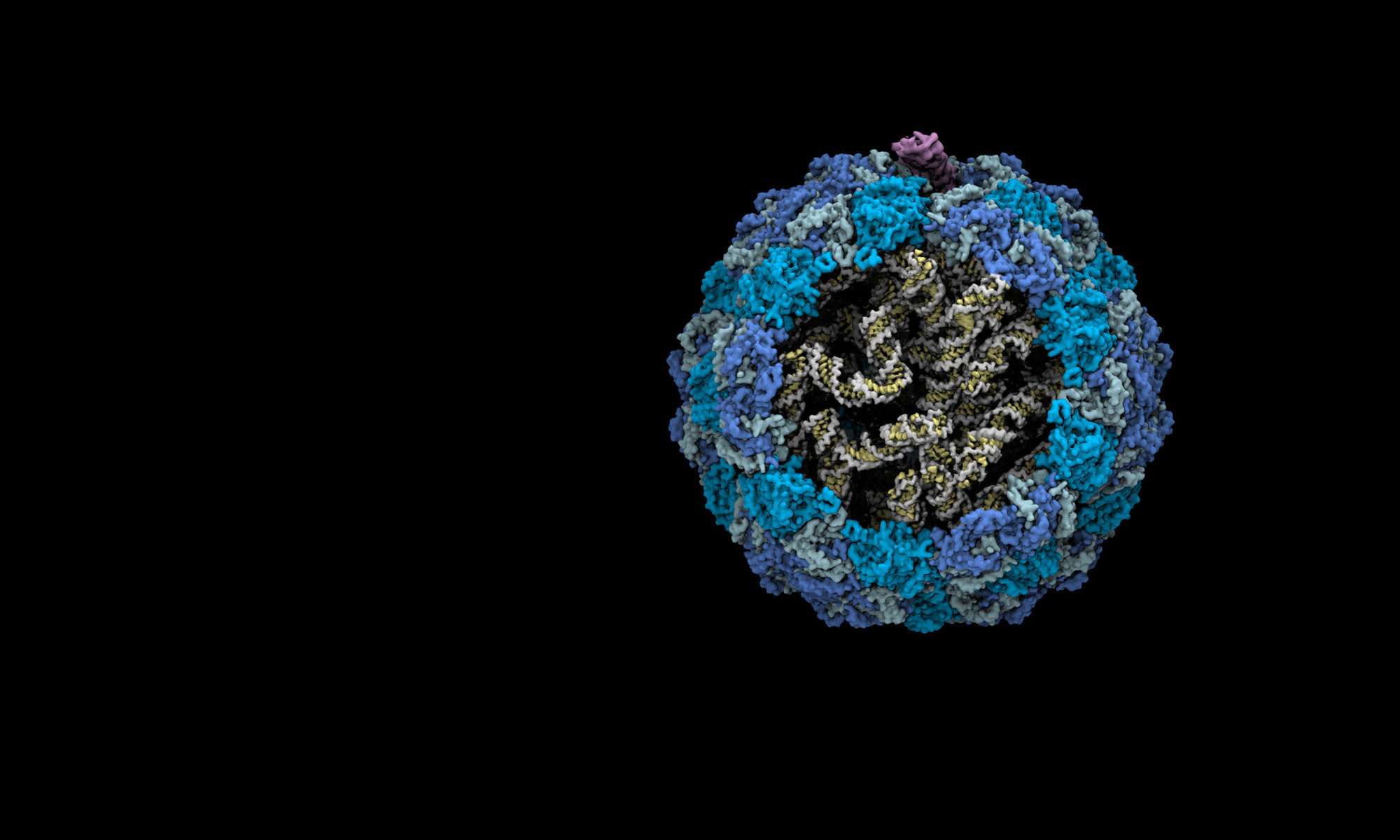
Thongchol, J.# , Yu, Z. # , Harb, L., Lin, Y., Koch, M., Theodore, M., Narsaria, U., Shaevitz, J., Gitai, Z., Wu, Y., Zhang, J. * , and Zeng, L. * , “Removal of Pseudomonas Type IV Pili by a Small RNA Virus “, Science . 2024 April 5; 384, doi :10.1126/science.adl0635. #, co-first author. * , co-corresponding author.
Meng R# , Xing Z# , Chang JY# , Yu Z, Thongchol J, Xiao W, Wang Y, Chamakura K, Zeng Z, Wang F, Young R, Zeng L, Zhang J. “Structural basis of Acinetobacter type IV pili targeting by an RNA virus “. Nat Commun . 2024 Mar 29;15(1):2746. doi: 10.1038/s41467-024-47119-5. PMID: 38553443. #, co-first author.
Thongchol J, Zhang J. “Purification of Single-Stranded RNA Bacteriophages and Host Receptors for Structural Determination Using Cryo-Electron Microscopy “. Methods Mol Biol . 2024;2793:185-204. doi: 10.1007/978-1-0716-3798-2_13. PMID: 38526732.
2023:
Zhang J, Lair C, Roubert C, Amaning K, Barrio MB, Benedetti Y, Cui Z, Xing Z, Li X, Franzblau SG, Baurin N, Bordon-Pallier F, Cantalloube C, Sans S, Silve S, Blanc I, Fraisse L, Rak A, Jenner LB, Yusupova G, Yusupov M, Zhang J, Kaneko T, Yang TJ, Fotouhi N, Nuermberger E, Tyagi S, Betoudji F, Upton A, Sacchettini JC, Lagrange S. “Discovery of natural-product-derived sequanamycins as potent oral anti-tuberculosis agents “. Cell . 2023 Feb 20:S0092-8674(23)00102-2. doi: 10.1016/j.cell.2023.01.043.
Thongchol J, Lill Z, Hoover Z, Zhang J. “Recent Advances in Structural Studies of Single-Stranded RNA Bacteriophages “. Viruses . 2023;15(10):1985. Published 2023 Sep 23. doi:10.3390/v15101985
2022:
Kustigian L, Gong X, Gai W, Thongchol J, Zhang J, Puchalla J, Carr CM, Rye HS. “GTP-stimulated membrane fission by the N-BAR protein AMPH-1 “. Traffic . 2022 Nov 26. doi: 10.1111/tra.12875.
Zeng Y, Jiang M, Robinson S, Peng Z, Chonira V, Simeon R, Tzipori S, Zhang J*, Chen Z*. “A Multi-Specific DARPin Potently Neutralizes Shiga Toxin 2 via Simultaneous Modulation of Both Toxin Subunits “. Bioengineering . 2022; 9(10):511. https://doi.org/10.3390/bioengineering9100511
Jiang M, Shin J, Simeon R, Chang J-Y, Meng R, Wang Y, Shinde O, Li P, Chen Z, Zhang J. “Structural dynamics of receptor recognition and pH-induced dissociation of full-length Clostridioides difficile Toxin B “. PLoS Biol 2022; 20(3): e3001589. https://doi.org/10.1371/journal.pbio.3001589
Chang J, Gorzelnik KV, Thongchol J, Zhang J. “Structural Assembly of Qβ Virion and Its Diverse Forms of Virus-Like Particles “. Viruses . 2022; 14(2):225. https://doi.org/10.3390/v14020225
Cui Z, Li X, Shin J, Gamper H, Hou YM, Sacchettini JC, Zhang J. “Interplay between an ATP-binding cassette F protein and the ribosome from Mycobacterium tuberculosis “. Nat Commun . 2022 Jan 21;13(1):432. doi: 10.1038/s41467-022-28078-1.
2021:
2020:
Brock DJ, Kondow-McConaghy H, Allen J, Brkljača Z, Kustigian L, Jiang M, Zhang J, Rye H, Vazdar M, Pellois JP. “Mechanism of cell penetration by permeabilization of late endosomes: interplay between a multivalent TAT peptide and Bis (monoacylglycero) phosphate ” .Cell chemical biology 27 (10), 1296-1307. e5.
Chang J, Cui Z, Yang K, Huang J, Minary P, Zhang J. “Hierarchical Natural Move Monte Carlo Refines Flexible RNA Structures into Cryo-EM Densities “. RNA . 2020 Aug 21:rna.071100.119. doi: 10.1261/rna.071100.119.
Dou T, Li Z, Zhang J, Evilevitch A, Kurouski D. “Nanoscale Structural Characterization of Individual Viral Particles Using Atomic Force Microscopy Infrared Spectroscopy (AFM-IR) and Tip-Enhanced Raman Spectroscopy (TERS) “. Anal Chem . 2020 Aug 18;92(16):11297-11304. doi: 10.1021/acs.analchem.0c01971. Epub 2020 Aug 4. PMID: 32683857.
Krieger IV, Kuznetsov V, Chang J, Zhang J, Moussa SH, Young RF, Sacchettini JS. “The Structural Basis of T4 Phage Lysis Control: DNA as the Signal for Lysis Inhibition “.J Mol Biol . 2020 Jul 24;432(16):4623-4636. doi: 10.1016/j.jmb.2020.06.013. Epub 2020 Jun 17.
2019:
Peng Z, Simeon R, Mitchell SB, Zhang J, Feng H, Chen Z. “Designed Ankyrin Repeat Protein (DARPin) Neutralizers of TcdB from Clostridium difficile Ribotype 027 “. mSphere . 2019 Oct 2;4(5). pii: e00596-19. doi: 10.1128/mSphere.00596-19.
Wang B, Zhang J, Wu Y. “A Multiscale Model for the Self-assembly of Coat Proteins in Bateriophage MS2 “. J Chem Inf Model . 2019 Aug 14. doi: 10.1021/acs.jcim.9b00514.
Meng R, Jiang M, Cui Z, Chang J, Yang K, Jakana J, Yu X, Wang Z, Hu B*, Zhang J*. “Structural basis for the adsorption of a single-stranded RNA bacteriophage “. Nature Communications . 2019 July 16;10, Article number: 3130 (2019)
Simeon R, Jiang M, Chamoun-Emanuelli AM, Yu H, Zhang Y, Meng R, Peng Z, Jakana J, Zhang J*, Feng H*, Chen Z*. “Selection and characterization of ultrahigh potency designed ankyrin repeat protein inhibitors of C. difficile toxin B “. PLoS Biol . 2019 Jun 24;17(6):e3000311. doi: 10.1371/journal.pbio.3000311.
2018:
2017:
Cui Z, Gorzelnik KV, Chang J, Langlais C, Jakana J, Young R, Zhang J. “Structures of Qβ virions, virus-like particles, and the Qβ–MurA complex reveal internal coat proteins and the mechanism of host lysis” . Proc Natl Acad Sci USA . 2017; doi:10.1073/pnas.1707102114
Yang K, Chang J, Cui Z, Li X, Meng R, Duan L, Thongchol J, Jakana J, Huwe CM, Sacchettini JC, Zhang J. “Structural insights into species-specific features of the ribosome from the human pathogen Mycobacterium tuberculosis” . Nucleic Acids Res. . 2017; doi:10.1093/nar/gkx785 (Cover Story )
Weaver J, Jiang M, Roth A, Puchalla J, Zhang J*, Rye HS*. “GroEL actively stimulates folding of the endogenous substrate protein PepQ” . Nat. Commun. . 2017; doi:10.1038/ncomms15934
2016:
Gorzelnik KV, Cui Z, Reed CA, Jakana J, Young R, Zhang J. “Asymmetric cryo-EM structure of the canonical Allolevivirus Qβ reveals a single maturation protein and the genomic ssRNA in situ” . Proc Natl Acad Sci USA . 2016; doi: 10.1073/pnas.1609482113 (Cover Story )
Lee JR, Xie X, Yang K, Zhang J, Lee SY, Shippen DE. “Dynamic interactions of Arabidopsis TEN1: stabilizing telomeres in response to heat stress.” . Plant Cell . 2016; doi: 10.1105/tpc.16.00408
Yang K, Ren Z, Raushel FM*, Zhang J*. “Structures of the Carbon-Phosphorus Lyase Complex Reveal the Binding Mode of the NBD-like PhnK.” . Structure . 2016; 24 (1): 37-42
2015:
Before 2015:
Zhang J, Minary P, Levitt M. “Multiscale natural moves refine macromolecules using single-particle electron microscopy projection images” . Proc Natl Acad Sci USA . 2012; 109 (25): 9845-50
Douglas NR, Reissmann S, Zhang J, Chen B, Jakana J, Kumar R, Chiu W, Frydman J. “Dual action of ATP hydrolysis couples lid closure to substrate release into the group II chaperonin chamber” .Cell . 2011; 144 (2): 240-52
Zhang J, Baker ML, Schröder GF, Douglas NR, Reissmann S, Jakana J, Dougherty M, Fu CJ, Levitt M, Ludtke SJ, Frydman J, Chiu W. “Mechanism of folding chamber closure in a group II chaperonin” . Nature . 2010; 463 (7279): 379-83
Zhang J, Chiu W. “Electron cryo-microscopy of molecular machines” . Encyclopedia of Biophysics (Springer). 2013;573-7
DiMaio F, Zhang J, Chiu W, Baker D. “Cryo-EM model validation using independent map reconstructions” . Protein Sci. . 2013;22(6):865-8
Zhang Q, Dai X, Cong Y, Zhang J , Chen DH, Dougherty MT, Wang J, Ludtke SJ, Schmid MF, Chiu W. “Cryo-EM structure of a molluscan hemocyanin suggests its allosteric mechanism” . Structure . 2013; 21 (4): 604-13
Zhang J, Ma B, DiMaio F, Douglas NR, Joachimiak LA, Baker D, Frydman J, Levitt M, Chiu W. “Cryo-EM structure of a group II chaperonin in the prehydrolysis ATP-bound state leading to lid closure” . Structure . 2011; 19 (5): 633-9
Knee KM, Goulet DR, Zhang J, Chen B, Chiu W, King JA. “The group II chaperonin Mm-Cpn binds and refolds human γD crystallin” . Protein Sci. . 2011; 20 (1): 30-41
Nakamura N, Shimizu Y, Shinkawa T, Nakata M, Bammes B, Zhang J, Chiu W. “Automated specimen search in cryo-TEM observation with DIFF-defocus imaging” . J Electron Microsc (Tokyo) . 2010; 59 (4): 299-310
Baker ML, Zhang J, Ludtke SJ, Chiu W. “Cryo-EM of macromolecular assemblies at near-atomic resolution” . Nat Protoc. . 2010; 5 (10): 1697-708
Pintilie GD, Zhang J, Goddard TD, Chiu W, Gossard DC. “Quantitative analysis of cryo-EM density map segmentation by watershed and scale-space filtering, and fitting of structures by alignment to regions” . J Struct Biol. . 2010; 170 (3): 427-38
Zhang J, Nakamura N, Shimizu Y, Liang N, Liu X, Jakana J, Marsh MP, Booth CR, Shinkawa T, Nakata M, Chiu W. “JADAS: a customizable automated data acquisition system and its application to ice-embedded single particles” . J Struct Biol. . 2009; 165 (1): 1-9 (Cover Story )
Pintilie G, Zhang J, Chiu W, Gossard D. “Identifying Components in 3D Density Maps of Protein Nanomachines by Multi-scale Segmentation” . IEEE NIH Life Sci Syst Appl Workshop . 2009: 2009 44-47
Chen DH, Luke K, Zhang J, Chiu W, Wittung-Stafshede P. “Location and flexibility of the unique C-terminal tail of Aquifex aeolicus co-chaperonin protein 10 as derived by cryo-electron microscopy and biophysical techniques” . J Mol Biol. . 2008; 381 (3): 707-17